(http://www.stats.ox.ac.uk/~macaulay/papers/richards_2000.pdf)
(http://www.hotep.bigstep.com/27082001.htm).
Graph taken from http://www.chitpavans.com/chitpavangenetics2.html
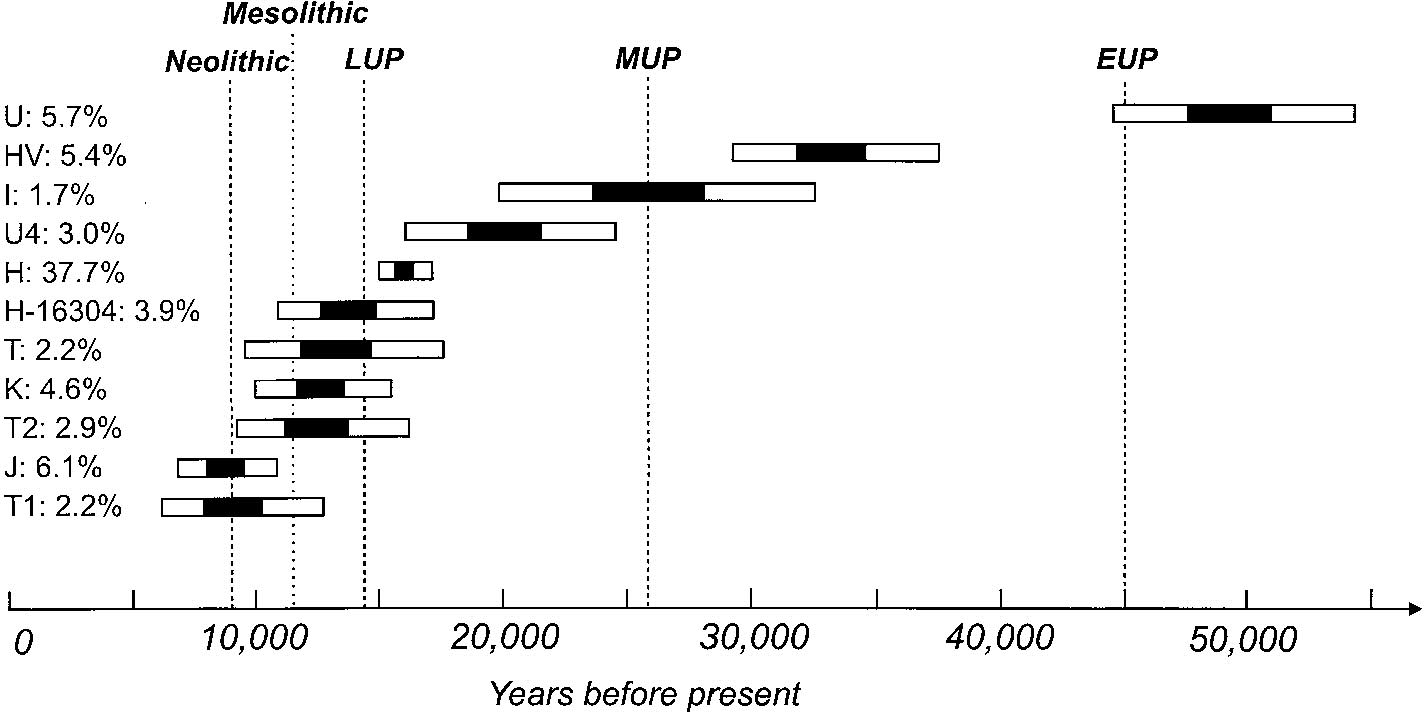
Note that the origin of haplogroup H (Helena) is Central and West Eurasia.
Human mitochondria (the energy-producing organelles outside the nucleus of cells) DNA is passed down generations only through females. Thus, testing of the DNA in mitochondria can aid in establishing matrilineal lines. (Establishing matrilineal lines with documents is more difficult than for patrilineal lines in most European lines because the surnames of females are lost in naming children; exceptions are Spanish and Portuguese.)
mtDNA is a circular structure composed of 16,569 nucleotides, also called base pairs. (It is believed that the mitochondria were originally parasitic bacteria which, through evolution, became symbiotic with the human cell and then became an integral part of the cell; bacterium DNA is circular.) There is a 400 base-pair section labeled 16001-16400 that evolves faster than other sections, so it is used to distinguish human groups that evolved separately. In the table below there are two sets of four rows that list the 400 base pairs: the first row starts at position 16001 and ends at 16100, the second row encompasses 16101-16200, the third row encompasses 16201-16300 and the fourth row encompasses 16301-16400. The Cambridge Reference Sequence (CRS) is the sequence of base pairs for an arbitrary person to which all other persons are compared.
Average number of mtDNA base pairs differences between two humans is about 10-66. The HVR1 test indicates to which of the major 36 World (10 for 99% of Europeans) haplogroups you belong. This could be millions of people! If it is a rare haplogroup, it could be a much smaller group. Mine is haplogroup I (Iris), in which are <2% of Europeans. This is most helpful for genealogy when you think you are related to someone through your maternal line. Test HVR1 for both to see if they are the same; if the same there is a 50% probability that you have a common ancestor 52 generations ago or less. For better resolution, do the HVR2 test; if still the same there is a 50% probability that you have a common ancestor 28 generations ago or less. So, it is not accurate enough for genealogy.
Full sequencing of mtDNA is equivalent to 41 markers on the Y-chromosome for genealogical use in the strict female line. It is difficult to follow a maternal line back many generations, since surnames usually change each generation.
I have determined the mtDNA pattern for my wife, Jeanne Muriel Baril, and her matrilineal line. Their earliest known maternal ancestor is Isabel MacEachern (b c1805 Scotland). The result is given in the table below in the second set of four rows of base pairs with the differences from the Cambridge Reference Sequence (CRS) in bold red. (There are four different base pairs labeled by Adenine, Cytosine, Guanine and Thymine.) In this case there are no differences; i.e., Jeanne has the same mtDNA as the Cambridge Reference Sequence. Another way to say is that the haplogroup H, to which Jeanne belongs, is the haplogroup taken as the Cambridge Reference Sequence.
| mtDNA HRV1 results for Jeanne Muriel Baril. (Starts at 16001, ends at 16569 = 569 base pairs.) | ||||||||||
| Cambridge Reference
Sequence (CRS): Jeanne Muriel Baril relative mutations (none): |
||||||||||
| 16001-10 | 16011-20 | 16021-30 | 16031-40 | 16041-50 | 16051-60 | 16061-70 | 16071-80 | 16081-90 | 16091-100 | Add |
| ATTCTAATTT | AAACTATTCT | CTGTTCTTTC | ATGGGGAAGC | AGATTTGGGT | ACCACCCAAG | TATTGACTCA | CCCATCAACA | ACCGCTATGT | ATTTCGTACA | +0 |
| TTACTGCCAG | CCACCATGAA | TATTGTACGG | TACCATAAAT | ACTTGACCAC | CTGTAGTACA | TAAAAACCCA | ATCCACATCA | AAACCCCCTC | CCCATGCTTA | +100 |
| CAAGCAAGTA | CAGCAATCAA | CCCTCAACTA | TCACACATCA | ACTGCAACTC | CAAAGCCACC | CCTCACCCAC | TAGGATACCA | ACAAACCTAC | CCACCCTTAA | +200 |
| CAGTACATAG | TACATAAAGC | CATTTACCGT | ACATAGCACA | TTACAGTCAA | ATCCCTTCTC | GTCCCCATGG | ATGACCCCCC | TCAGATAGGG | GTCCCTTGAC | +300 |
| CACCATCCTC | CGTGAAATCA | ATATCCCGCA | CAAGAGTGCT | ACTCTCCTCG | CTCCGGGCCC | ATAACACTTG | GGGGTAGCTA | AAGTGAACTG | TATCCGACAT | +400 |
| CTGGTTCCTA | CTTCAGGGTC | ATAAAGCCTA | AATAGCCCAC | ACGTTCCCCT | TAAATAAGAC | ATCACGATG | +500 | |||
| Age ranges for some founding haplogroups (http://www.stats.ox.ac.uk/~macaulay/papers/richards_2000.pdf) (http://www.hotep.bigstep.com/27082001.htm). |
 |
| Note that, in this study, haplogroup H, where Jeanne's mtDNA lies, is 38% of the European population in this study and originated c16,000 ybp. |
 Graph taken from http://www.chitpavans.com/chitpavangenetics2.html Note that the origin of haplogroup H (Helena) is Central and West Eurasia. |
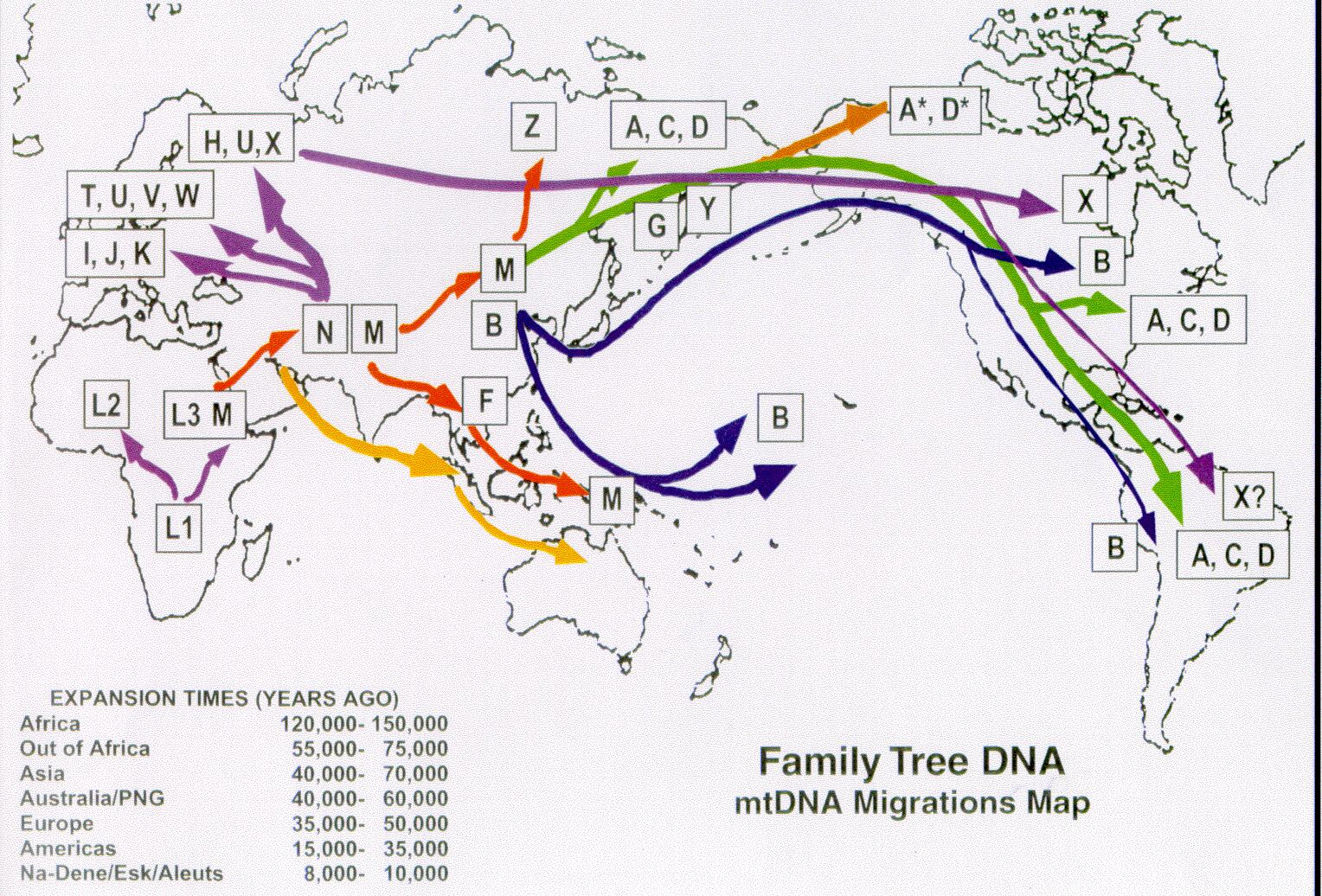
| Migration map for founding haplogroups, courtesy of
Family Tree DNA Note that haplogroup ... originated in ... area. |
 |
The major haplogroups for Europeans are H, J, U, T, K, X, V and I. (See Bryan Syke's book The Seven Daughters of Eve.)
| haplogroup | % of Europeans | years before present | origin |
| H (Helena) | 47 | 20,000 | South France |
| J (Jasmine) | 17 | 10,000 | Middle East |
| U (Ursula) | 11 | 45,000 | Greece |
| T (Tara) | 9 | 17,000 | Tuscany |
| K (Katrina) | 6 | 15,000 | North Italy |
| X (Xenia) | 6 | 25,000 | Georgia, Asia |
| V (Velda) | 5 | 17,000 | North Spain |
| I (Iris) | 2 | 26,000 | ? |
For a more recent excellent discourse on mtDNA compared to Y-chromosome tests, see Bryan Sykes, Adam's Curse: A Future Without Men.
View all known female descendants (and their husbands) of Isabel MacEachern (b c1805 Scotland).
Roper
Genetics Project
Franklin Genetics
Project
Little/Klein/Cline Genetics
Project