Y-Chromosome Biallelic Haplogroups
L. David Roper (roperld@vt.edu)
(www.roperld.com)
Y-Chromosome Haplogroup Trees
Y-chromosome haplogroup tree
The Journey of Man, A Genetic Odyssey by Spencer
Wells, p. 182.
Information about the
Y-chromosome haplogroups.
Y-DNA Haplogroup Tree 2007
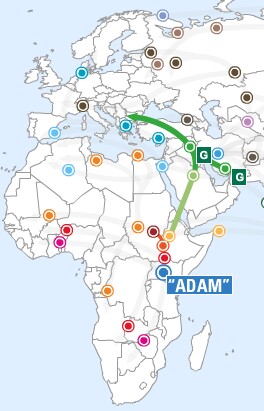
Migration Routes
Biallelic SNP markers are single base-pair mutations (polymorphisms) that occur at different Y-chromosome
locations about once every 7000 years. SNP=Single Nucleotide Polymorphism.
There are over 150 known haplogroups. Haplotypes defined by the 31 STR
markers are subgroups under the haplogroups
HAPLOGROUP
DEFINITIONS: Family
Tree DNA provided the following thumbnail summaries of the different
haplogroups :
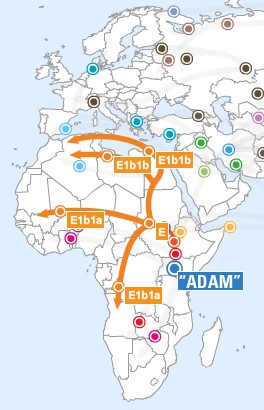
- Haplogroup A is found mainly in the Southern Nile region and Southern Africa. It represents the oldest and most diverse of the human Y-chromosome haplogroups. It is believed to be the haplogroup corresponding to Y-chromosomal Adam.
- Haplogroup B is one of the oldest Y-chromosome lineages in
humans. Haplogroup B is found exclusively in Africa. This lineage was the first
to disperse around Africa. There is current archaeological evidence supporting
a major population expansion in Africa approximately 90-130 thousand years ago.
It has been proposed that this event may have spread Haplogroup B throughout
Africa. Haplogroup B appears at low frequency all around Africa, but is at its
highest frequency in Pygmy populations.
- Haplogroup C is found throughout mainland Asia, the south
Pacific, and at low frequency in Native American populations. Haplogroup C
originated in southern Asia and spread in all directions. This lineage
colonized New Guinea, Australia, and north Asia, and currently is found with
its highest diversity in populations of India.
- Haplogroup C3 is believed to have originated in southeast or
central Asia. This lineage then spread into northern Asia, and then into the
Americas.
- Haplogroup D is an old lineage that evolved in Asia where it is hypothesized to have been widely distributed. This Haplogroup was present in the first people to colonize Japan. This Haplogroup was later displaced from much of Asia by other colonizing groups, but is still present at intermediate frequencies in the aboriginal Japanese and on the Tibetan plateau. It is also found at low frequencies in Mongolian populations and the Altais people of central Asia.
- Haplogroup D1 lineage is a descendent of the D lineage and is currently present in Southeast Asia and Tibet. Like its progenitor it is also found in low frequencies in Mongolian populations, but unlike D it is completely absent from Japan.
- Haplogroup D2 most likely derived from the D lineage in Japan.
It is completely restricted to Japan, and is a very diverse lineage within the
aboriginal Japanese and in the Japanese population around Okinawa.
- Haplogroup E probably arose in Northeast Africa, if one looks only at the
concentration and variety of E subclades in that area today. But the fact that Haplogroup E is
closely linked with Haplogroup D, which is not found in Africa, leaves open the possibility that
E first arose in the Near or Middle East and was subsequently carried into Africa by a back
migration
- Haplogroup E3a is an Africa lineage. It is currently
hypothesized that this haplogroup dispersed south from northern Africa within
the last 3,000 years, by the Bantu agricultural expansion. E3a is also the most
common lineage among African Americans.
- Haplogroup E3b is believed to have evolved in the Middle East.
It expanded into the Mediterranean during the Pleistocene Neolithic expansion.
It is currently distributed around the Mediterranean, southern Europe, and in
north and east Africa.
- Haplogroup F is the parent of all Y-DNA haplogroups G through R and contains more than
90% of the world’s population. Haplogroup F was in the original migration out of Africa, or else it was founded soon afterward, because F and its sub-haplogroups are primarily found outside, with very few inside, sub-Saharan Africa. The founder of F could have lived between 60,000 and 80,000 years ago, depending on the time of the out-of-Africa migration.
- Haplogroup G may have originated in India or Pakistan, and has
dispersed into central Asia, Europe, and the Middle East. The G2 branch of this
lineage (containing the P15 mutation) is found most often in Europe and the
Middle East.
- Haplogroup H is nearly completely restricted to India, Sri
Lanka, and Pakistan.
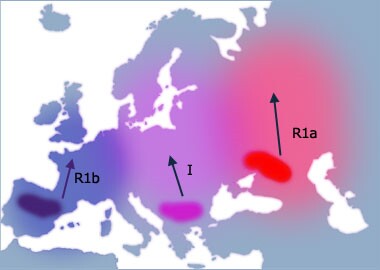
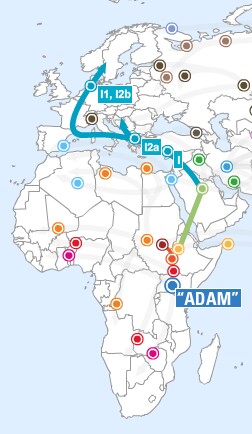
- Haplogroup I is a European haplogroup, representing nearly one-fifth of the
population. It is almost non-existent outside of Europe, suggesting that it arose in Europe.
Estimates of the age of haplogroup I suggest that it arose prior to the last Glacial Maximum.
- Haplogroup I1-M253 et al has highest frequency in Scandinavia, Iceland, and northwest Europe. In Britain, this haplogroup is often used as a marker for "invaders," Viking or Anglo-Saxon. The I1b-M227 subclade is concentrated in eastern Europe and the Balkans and appears to have arisen in the last one thousand to five thousand years. It has been reported in Germany, Czech Republic, Poland, Estonia, Ukraine, Switzerland, Slovenia, Bosnia, Macedonia, Croatia, and Lebanon.
- Haplogroup I2-M438 et al includes I2* which shows some membership from Armenia, Georgia and Turkey; I2a-P37.2, which is the most common form in the Balkans and Sardinia; I2a2-M26 is especially prevalent in Sardinia. I2b-M436 et al reaches its highest frequency along the northwest coast of continental Europe. I2b1-M223 et al occurs in Britain and northwest continental Europe. I2b1a-M284 occurs almost exclusively in Britain, so it apparently originated there and has probably been present for thousands of years.
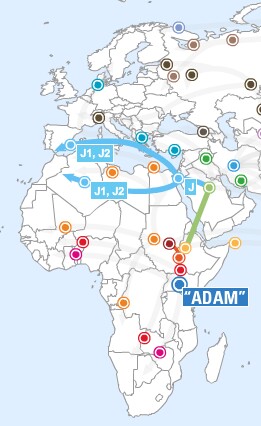
- Haplogroup J (J1) is found at highest
frequencies in Middle Eastern and north African populations where it most
likely evolved. This marker has been carried by Middle Eastern traders into
Europe, central Asia, India, and Pakistan.
- Haplogroup J2 originated in the northern portion of the
Fertile Crescent where it later spread throughout central Asia, the
Mediterranean, and south into India. As with other populations with
Mediterranean ancestry this lineage is found within Jewish populations. The
Cohen modal lineage is found in Haplogroup J2.
- Haplogroup K first appeared approximately 40,000
years ago in Iran
or southern
Central Asia.. See
http://en.wikipedia.org/wiki/Haplogroup_K_%28Y-DNA%29
.
- Haplogroup L is found primarily in India and Sri
Lanka, and has also spread into several Middle Eastern populations (Turks,
Saudis, and Pakistanis).
- Haplogroup M reaches its known peak in Papua New Guinea, totaling one-third to
two-thirds of population.
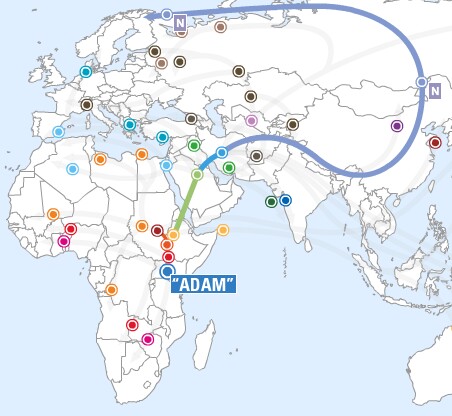
- Haplogroup N is distributed throughout Northern
Eurasia. It is the most common Y-chromosome type in Uralic speakers (Finns and
Native Siberian). This lineage most likely originated in northern China or
Mongolia and then spread into Siberia where it became a very common line in
western Siberia.
- Haplogroup O lineage dates to about 35,000 years ago
when it evolved in Central or East Asia. Over 80% of today's East Asian
population is haplogroup O, and this lineage is nearly completely restricted to
East Asian populations.
- Haplogroup O1 is found at very high frequency in the
aboriginal Taiwanese (possibly due to genetic drift). This haplogroup probably
originated in East Asia and later migrated into the south Pacific. Individuals
carrying this lineage are thought to have been important in the expansion of
the Austronesian language group into Taiwan, Indonesia, Melanesia, Micronesia,
and Polynesia.
- Haplogroup O2 has two primary lines, the 465 line
and the M95 line. Both lines are found in Asia. The 465 line is at high
frequency in Japanese and Korean populations and at low frequency in east Asia.
The M95 line is found in Southeast Asian populations (Malaysia, Vietnam,
Indonesia, and southern China).
- Haplogroup P is rarely found as an undifferentiated haplogroup. It is ancestral to
both haplogroup Q and the much larger haplogroup R. Origin in central Asia or the Altai region
of Siberia is suspected. Today the haplogroup is found at low frequencies in India, Pakistan,
and central Asia.
- Haplogroup Q is the lineage that links Asia and the Americas.
This lineage is found in North and Central Asian populations as well as native
Americans. This lineage is believed to have originated in Central Asia and
migrated through the Altai/Baikal region of northern Eurasia into the
Americas.
- Haplogroup Q3 is the only lineage strictly associated with
native American populations. This haplogroup is defined by the presence of the
M3 mutation (also known as SY103). This mutation occurred on the Q lineage 8-12
thousand years ago as the migration into the Americas was underway. There is
some debate as to on which side of the Bering Strait this mutation occurred,
but it definitely happened in the ancestors of the Native American
peoples.
- Haplogroup R is mainly represented in two lineages. Lineage R1a is thought to have
originated in the Eurasian Steppes north of the Black and Caspian Seas. It is associated with
the Kurgan culture, known for the domestication of the horse (approximately 3000 B.C.E.). This
lineage is currently found in central and western Asia, India, and in Slavic populations of
Eastern Europe. A well-known individual of the R1a lineage is Somerled founder of Clan Donald.
Lineage R1b originated prior to the end of the last ice age where it was concentrated in refugia
in southern Europe and Iberia and is the most common in European populations. It is especially
common in the west of Ireland where it approaches 100% of the population. This haplogroup contains
the Atlantic modal STR haplotype.
- Haplogroup R1a is believed to have originated in the Eurasian
Steppes north of the Black and Caspian Seas. This lineage is believed to have
originated in a population of the Kurgan culture, known for the domestication
of the horse (approximately 3000 B.C.E.). These people were also believed to be
the first speakers of the Indo-European language group. This lineage is
currently found in central and western Asia, India, and in Slavic populations
of Eastern Europe.
- Haplogroup R1b is the most common haplogroup in European
populations. It is believed to have expanded throughout Europe as humans
re-colonized after the last glacial maximum 10-12 thousand years ago. This
lineage is also the haplogroup containing the Atlantic modal haplotype
(HG1).
- Haplogroup R1b1 is a subgroup of R1b.
R1b1c is a subgroup of R1b1.
- Haplogroup R1b1c3 is arrived in eastern Britain before the Younger Dryas (13,000 -> 11,500 years ago) and re-expanded into Eastern Britain immediately after the YD. It is found in only 1% of the British sample population. ... Two-thirds of R1b1c3 representatives are found in eastern England, the Channel Islands and Dorchester, while 82% are restricted to eastern England: Norfolk, Southwell, Bourne and York. ... The distribution of the founder type suggest R1b1c3 may have mutated from R1b1c somewhere in the English Channel (when it was mostly dry), and from there moved up into Norfolk and north-east England. ... Norfolk and East Anglia were still linked to the Continent at this stage.
(The Origins of the British: A Genetic Detective Story
by Stephen Oppenheimer, p.154.)
- Haplogroup R1b1c6 is found primarily in the British Isles, but can also be found at lower frequencies around Western Europe.
- Haplogroup R1b1c7 is primarily found in Northern Ireland and contains the Niall Modal Haplotype.
- Haplogroup S is commonly found among populations of the highlands of Papua New Guinea. It is also found at lower frequencies in adjacent parts of Indonesia and
Melanesia.
- Haplogroup T is present at a low level throughout
Africa, Southwest Asia and Southern Europe. (Thomas Jefferson is in T. See
http://news.bbc.co.uk/2/hi/science/nature/6332545.stm).
R1b1 Haplogroup:
Y Biallelic SNP Marker |
Years Before Present |
Migration Route |
| M94 |
? |
In Africa |
| M168 |
50,000 |
Africa->Middle East |
| F(M89) |
45,000 |
Middle East->South West Asia |
| K(M9) |
40,000 |
South West Asia->North Central Asia |
| P(M45) |
35,000 |
North Central Asia->North West Asia |
| R(M207) |
? |
In North West Asia |
| 1(M173) |
30,000 |
North West Asia->Europe |
| b(M343) |
? |
In Europe |
| 1(P25) |
|
|
| a(M18), b(M73), c(M269), d(M335) |
|
|
| c1(M37), c2(M65), c3(M126), c4(M153), c5(M160), c6(M167), c7(M222), c8(P66), c9(S21), c10(S28) |
|
|
The Journey of Man, A Genetic Odyssey by Spencer
Wells
R1a Haplogroup:
| Y Biallelic SNP Marker |
Years Before Present |
Migration Route |
| M94 |
? |
In Africa |
| M168 |
50,000 |
Africa->Middle East |
| F(M89) |
45,000 |
Middle East->South West Asia |
| K(M9) |
40,000 |
South West Asia->North Central Asia |
| P(M45) |
35,000 |
North Central Asia->North West Asia |
| R(M207) |
? |
In North West Asia |
| 1(M173) |
30,000 |
North West Asia->Europe |
| a |
? |
In Europe |
E3b Haplogroup:
| Y Biallelic SNP Marker |
Years Before Present |
Migration Route |
| M94 |
? |
In Africa |
| M168 |
50,000 |
Africa->Middle East |
| M145 |
? |
? |
| E(M96) |
? |
? |
| 3(P2) |
? |
? |
| b(M35) |
? |
? |
G Haplogroup:
| Y Biallelic SNP Marker |
Years Before Present |
Migration Route |
| M94 |
? |
In Africa |
| M168 |
50,000 |
Africa->Middle East |
| F(M89) |
45,000 |
Middle East->South West Asia |
| G(M201) |
? |
? |
I Haplogroup:
| Y Biallelic SNP Marker |
Years Before Present |
Migration Route |
| M94 |
? |
In Africa |
| M168 |
50,000 |
Africa->Middle East |
| F(M89) |
45,000 |
Middle East->South West Asia |
| I(M170) |
? |
? |
I1b Haplogroup:
| Y Biallelic SNP Marker |
Years Before Present |
Migration Route |
| M94 |
? |
In Africa |
| M168 |
50,000 |
Africa->Middle East |
| F(M89) |
45,000 |
Middle East->South West Asia |
| I(M170) |
? |
? |
| 1(P38) |
? |
? |
| b(P37b) |
? |
? |
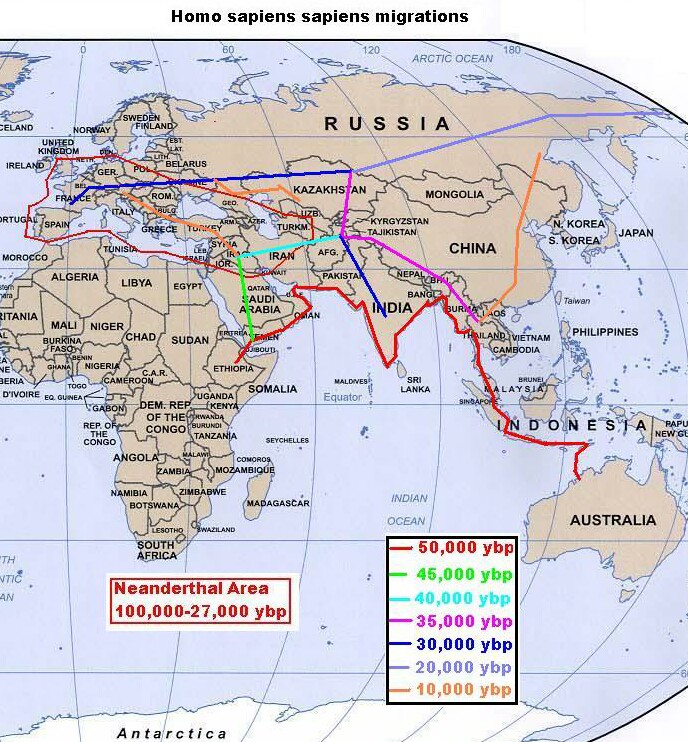
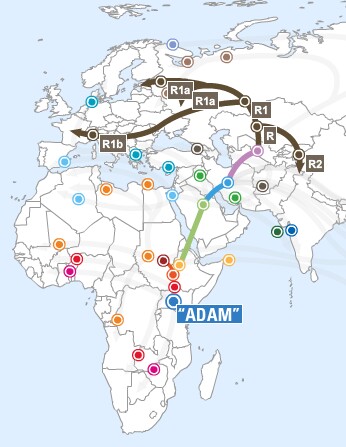
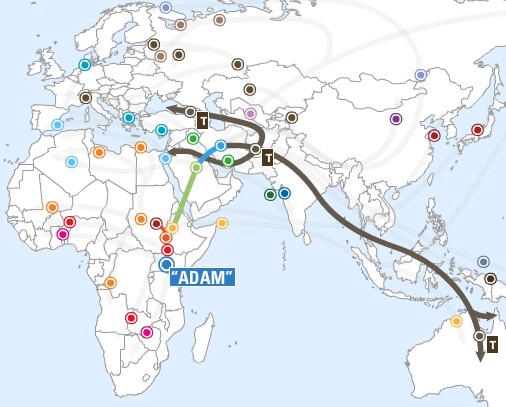
Probable human migrations out of Africa |
|
|
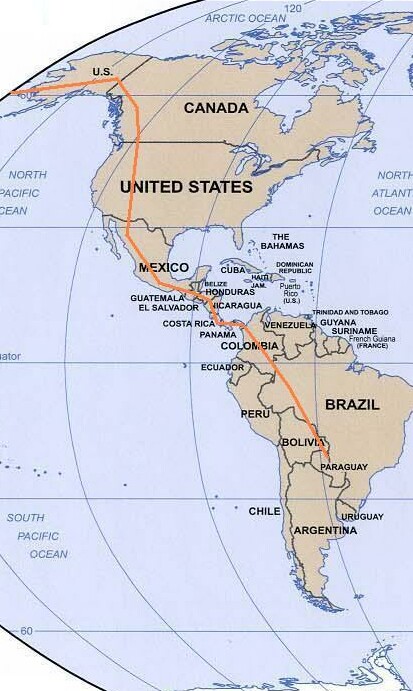
Migrations of humans out of refugia after the
Last Major Ice Age (~12,000 ybp) |
|
 |
See
http://www.scs.uiuc.edu/~mcdonald/WorldHaplogroupsMaps.pdf
for Y-chromosome and mtDNA haplogroups maps.
Migration routes: