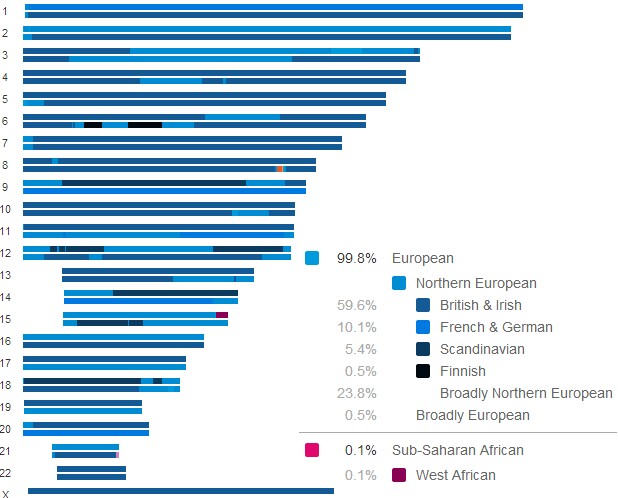
British & Irish |
59.6% |
|---|---|
French & German |
10.1% |
Scandinavian |
5.4% |
Finnish |
0.5 |
West African |
0.1 |

2.9% 80th percentile(Average 23andMe user: 2.7%)
Neanderthal Man: In Search of Lost Genomes

Introduction
Haplogroup I appears to have originated in the Near East or Caucasus Mountains about 40,000 years ago, not long after humans began migrating beyond Africa. Today the haplogroup is widespread in Europe, where it is present at levels of about 2%. Though fairly rare in the Near East, the haplogroup is found in a swath from the Caucasus region to Pakistan. Haplogroup I has also made relatively recent incursions into Africa, where it is found at levels of about 1% in Ethiopia and Egypt.
Into EuropeHaplogroup I began spreading into Europe just before the peak of the Ice Age, about 20,000 years ago. At first it was probably restricted to southeastern Europe, where conditions were somewhat warmer than in the frigid interior of the continent. After the glaciers retreated, the haplogroup drifted throughout much of Europe. Traces of that expansion can be seen today in the presence of haplogroup I among formerly Celtic-speaking populations in Scotland, Wales, Cornwall and Brittany in northern France.
Some Icelanders also bear haplogroup I, a suggestion that their ancestry traces back to British and Irish women who were taken away from their homes by Viking raiders during the 9th and 10th centuries AD.
But haplogroup I also appears to have been present in Scandinavia during the Viking era. Ancient DNA belonging to the haplogroup was extracted from skeletal remains at an early Christian cemetery in Denmark.
Today, haplogroup I is very rare in Denmark. But it is found at a level of about 4% in Finland and 2% in other parts of Scandinavia.
The Agricultural Revolution
While many other haplogroups show evidence of having moved from the Near East into Europe with the expansion of agriculture, I seems to have moved in the opposite direction. It appears to have moved from the Caucasus region south around the Caspian Sea and into southwestern Asia about 8,000 years ago, about the same time that agriculture spread throughout the region.
Today haplogroup I is distributed from the Caucasus to southern Pakistan, where it is most common among the Sindhi, a population that has been associated linguistically with the spread of farming. The haplogroup's present-day distribution appears to trace a migration along the northern corridor of southwestern Asia, because it does not extend southward into the Persian Gulf region, or as far as India.
Haplogroup I1Haplogroup I1 has been found at very low frequencies across central Asia. People bearing this haplogroup probably made their way west across Asia following the Silk Road, a corridor of trade that brought spices and silks to Europe, where I1 can be found at very low levels in Hungary and France.
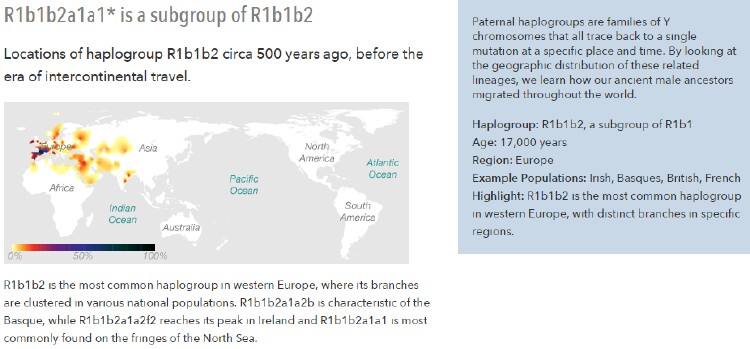
Haplogroup R is a widespread and diverse branch of the Y-chromosome tree that is extremely common in Europe, where it spread after the end of the Ice Age about 12,000 years ago. The haplogroup appears to have originated in southwestern Asia about 30,000 years ago. It then split into two main branches. R1 ultimately spread widely across Eurasia, from Iceland to Japan, whereas R2 mostly remained near its region of origin. Today it can be found in southwestern Asia and India.
Because of recent immigration, both branches of R are now found worldwide among men of European, Middle Eastern and South Asian descent – though our haplogroup maps indicate only their pre-colonial distributions.
R1 is the dominant haplogroup in Europe today, accounting for well over half of all men. After being confined to the continent's southern fringes during the Ice Age, it expanded rapidly in the wake of the receding glaciers about 12,000 years ago. Various branches of R1 also trace the many migrations that have shaped Europe since then, from the arrival of farmers between about 10,000 and 7,000 years ago to the movements of ethnic groups such as the Anglo-Saxons and Vikings.
Haplogroup R1bHaplogroup R1b was confined during the Ice Age to pockets of territory in Mediterranean Europe. The largest was in the Iberian peninsula and southern France, where men bearing the haplogroup created the famous cave paintings at Lascaux and Altamira. They also hunted mammoth, bison and other large game in a climate that was more like present-day Siberia's than the mild conditions prevailing in southern Europe today.
Some men bearing R1b Y-chromosomes also seem to have spent the Ice Age in the Balkans and Anatolia, where the haplogroup is still present today.
After the Ice Age, the haplogroup expanded rapidly in the wake of the retreating glaciers. Today R1b is by far the most common haplogroup in the western half of the continent.
Haplogroup R1b1b2R1b1b2 is the most common haplogroup in western Europe, where it is found in more than 50% of men. Ancient representatives of the haplogroup were among the first people to repopulate the western part of Europe after the Ice Age ended about 12,000 years ago. In the process the haplogroup differentiated into even more distinct groups that can trace the details of the post-Ice Age migrations.
Haplogroup R1b1b2a1a2bR1b1b2a1a2b arose about 20,000 years ago, when the Ice Age was at its peak. It appears to have originated among the ancestors of the present-day Basque, because of the relatively high diversity of the haplogroup in that population compared to neighboring ones. Today R1b1b2a1a2b is found in about 5% of Basque and 1% of Iberians.
Haplogroup R1b1b2a1a2f2R1b1b2a1a2f2 reaches its peak in Ireland, where the vast majority of men carry Y-chromosomes belonging to the haplogroup. Researchers have recently discovered that a large subset of men assigned to the haplogroup may be direct male descendants of an Irish king who ruled during the 4th and early 5th centuries. According to Irish history, a king named Niall of the Nine Hostages established the Ui Neill dynasty that ruled the island country for the next millennium.
Northwestern Ireland is said to have been the core of Niall's kingdom; and that is exactly where men bearing the genetic signature associated with him are most common. About 17% of men in northwestern Ireland have Y-chromosomes that are exact matches to the signature, and another few percent vary from it only slightly. In New York City, a magnet for Irish immigrants during the 19th and early 20th century, 2% of men have Y-chromosomes matching the Ui Neill signature. Genetic analysis suggests that all these men share a common ancestor who lived about 1,700 years ago. Among men living in northwestern Ireland today that date is closer to 1,000 years ago. Those dates neatly bracket the era when Niall is supposed to have reigned.
Outside Ireland, R1b1b2a1a2f2 is relatively common only along the west coast of Britain.
Haplogroup R1b1b2a1a1Today R1b1b2a1a1 is found mostly on the fringes of the North Sea in England, Germany and the Netherlands, where it reaches levels of one-third. That distribution suggests that some of the first men to bear the haplogroup in their Y-chromosomes were residents of Doggerland, a real-life Atlantis that was swallowed up by rising seas in the millennia following the Ice Age.
Doggerland was a low-lying region of forests and wetlands that must have been rich in game; today, fishing trawlers in the North Sea occasionally dredge up the bones and tusks of the mastodons that roamed there. Doggerland had its heyday between about 12,000 years ago, when the Ice Age climate began to ameliorate, and 9,000 years ago, when the meltwaters of the gradually retreating glaciers caused sea levels to rise, drowning the hunter's paradise. Doggerland's inhabitants retreated to the higher ground that is now the North Sea coast.
Name |
Risk (%) |
Avg. Risk (%) |
Compared to Avg. |
|---|---|---|---|
Atrial Fibrillation |
33.9 |
27.2 |
1.25x |
Age-related Macular Degeneration |
24.6 |
6.5 |
3.76x |
Rheumatoid Arthritis |
6.1 |
2.4 |
2.56x |
Chronic Kidney Disease |
4.2 |
3.4 |
3.4x |
Restless Leg Syndrome |
2.5 |
2.0 |
1.25x |
Exfoliation Glaucoma |
2.2 |
0.7 |
2.90x |
Parkinson's Disease |
2.0 |
1.6 |
1.23x |
Type 1 Diabetes |
1.5 |
1.0 |
1.43x |
Esophageal Squamous Cell Carcinoma |
0.43 |
0.36 |
1.21x |
Stomach Cancer |
0.28 |
0.23 |
1.22x |
Scleroderma |
0.08 |
0.07 |
1.24x |
Name |
Risk (%) |
Avg. Risk (%) |
Compared to Avg. |
|---|---|---|---|
Type 2 Diabetes |
19.6 |
25.7 |
0.76x |
Gout |
17.1 |
22.8 |
0.75x |
Venous Thromboembolism |
9.0 |
12.3 |
0.73x |
Prostate Cancer |
8.7 |
17.8 |
0.49x |
Psoriasis |
5.8 |
11.4 |
0.51x |
Alzheimer's Disease |
4.3 |
7.2 |
0.60x |
Melanoma |
2.2 |
2.9 |
0.75x |
Multiple Sclerous |
0.20 |
0.34 |
0.59x |
Crohn's Disease |
0.14 |
0.53 |
0.27x |
Celiac Disease |
0.06 |
0.12 |
0.48x |
Primary Biliary Cirrhosis |
0.04 |
0.08 |
0.48x |
Name |
Risk (%) |
Avg. Risk (%) |
Compared to Avg. |
|---|---|---|---|
Obesity |
73.9 |
63.9 |
1.16x |
Coronary Heart Disease |
48.9 |
46.8 |
1.04x |
Lung Cancer |
8.2 |
8.5 |
0.97x |
Gallstones |
6.2 |
7.0 |
0.88x |
Colorectal Cancer |
5.6 |
15.6 |
1.00x |
Ulcerative Colitis |
0.63 |
0.77 |
0.83x |
Bipolar Disorder |
0.10 |
0.10 |
1.00x |
Breast Cancer |
0.00 |
0.00 |
1.00x |
Lupus |
0.00 |
0.530.00 |
1.00x |
Cystic Fibrosis: Has one or more genetic variants tested for this condition.